The Atlantic reports on Phylo, an online flash video game that has helped solve a host of DNA problems and greatly expanded our genetic understanding of a variety od diseases, such as diabetes, cancer, and Alzheimers.
As the Phylo website explains:
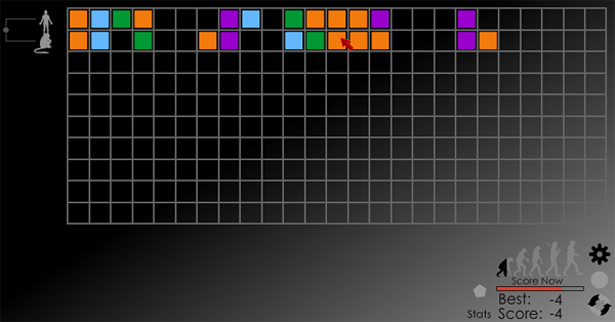
A sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity. These similarities may be consequences of functional, structural, or evolutionary relationships between the sequences. From such an alignment, biologists may infer shared evolutionary origins, identify functionally important sites, and illustrate mutation events. More importantly, biologists can trace the source of certain genetic diseases.
Traditionally, multiple sequence alignment algorithms use computationally complex heuristics to align the sequences. Unfortunately, the use of heuristics do not guarantee global optimization as it would be prohibitively computationally expensive to achieve an optimal alignment….
Humans have evolved to recognize patterns and solve visual problems efficiently. By abstracting multiple sequence alignment to manipulating patterns consisting of colored shapes, we have adapted the problem to benefit from human capabilities. By taking data which has already been aligned by a heuristic algorithm, we allow the user to optimize where the algorithm may have failed.
Through use of this gamification model, in only a year online and with 17,000 registered users, the researchers have received more than 350,000 solutions to sequence alignment problems. To tap into the incredible power of crowdsourcing, these researchers developed an engaging way to create the kind of data they were seeking and then made it available to a wide online gaming community. Rather than alignment solutions being achieved by “prohibitively expensive” computational methods, it makes perfect sense to harness human ingenuity and our efficient problem-solving capabilities to this monstrous research challenge, and others like it.
Like art.